In the laboratory that witnessed the dawn of the molecular biology revolution seven decades ago, scientists are creating new kinds of life, and accelerating the pace of evolution.
Proteins are the building blocks of all living things and the aim of their research is to create synthetic proteins far beyond the complexity of anything currently made by chemists, from proteins that can be used as novel drugs, such as antibiotics, or that can enhance biomaterials, build molecular machines, create wonder materials, or increase the ability of organisms to absorb greenhouse gases.

The work by Jason Chin’s team at the Medical Research Council’s Laboratory of Molecular Biology, LMB, in Cambridge, where many a Nobel prize has been won, shows sufficient commercial promise that a company, Constructive Bio was set up south of Cambridge in Whittlesford Parkway with $15 million to develop his tools. The slogan of his company is ‘Life, reimagined.’
‘These approaches allow us to turn cells into programmable factories for making whole new classes of molecules, with potential applications in diverse sectors,’ he said.
BIRTHPLACE OF MOLECULAR BIOLOGY
The LMB is famous for launching the revolution in molecular biology in the 1950s-60s, when physics – in the form of a technique called X ray diffraction – collided with biology to reveal the extraordinary molecular machinery of life, such as DNA, which stores the recipe to make proteins.
Many pioneering models of the molecules of life created there can be seen in the Science Museum, from Watson and Crick’s DNA double helix model to an extraordinary representation of the muscle protein myosin, suspended on a forest of rods.
In recent years the Chin Lab, the largest at the LMB, has pioneered the development of methods for reprogramming the DNA code of living organisms, rewriting the near-universal genetic code of natural life, focusing their efforts on a workhorse of molecular biology, the gut bacterium E. coli, which is also used by biotechnologists to make useful proteins.

All living things on Earth, from humans to E. coli, are built from proteins that are created from a set of 20 chemical units, called the canonical amino acids. Four years ago, Jason Chin’s laboratory reported that it could make proteins composed of new amino acids beyond those ‘canonical 20’.
But though ‘non canonical’, these building blocks were limited to being only variants of what are called alpha-L-amino acids. Because the many-faceted cellular machinery that strings together amino acids into proteins has evolved over billions of years, they ran into what Jason Chin called an ‘evolutionary deadlock’ when trying to introduce new types of building blocks – changing one cellular component had knock on effects such that, if the new component was to work, it demanded changes in other components.

EXTENDING THE ALPHABET OF LIFE
To overcome ‘this chicken or egg problem’, as Chin put it, Daniel Dunkelmann, Carlos Piedrafita and colleagues in his laboratory reported in the journal Nature this month that they have found a way to add new classes of building blocks to the genetic code in living organisms.
His team had previously added hundreds of ‘non-canonical’ alpha-L-amino acids to the genetic code by redesigning key enzymes, called synthetases. These work with transfer RNA, or tRNA, which carries amino acids to the ribosome, a complex molecule of around half a million atoms. The ribosome turns the genetic code of an organism into proteins, cellular building blocks that themselves are tangled strings of amino acids, by assembling the amino acids carried by each tRNA.
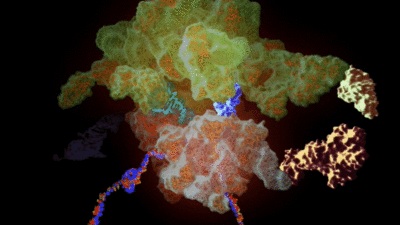
He calls the new synthetases ‘orthogonal’ synthetases, by which he means they work alongside the ones that naturally operate in a cell, without interfering with them, using orthogonal versions of transfer RNA.
In the work unveiled this month, Chin developed orthogonal synthetases that work with eight new, non-alpha-L-amino acid, building blocks, and showed that several of these new building blocks could be smuggled through the ribosome to make proteins containing new classes of building blocks.
‘We have been trying for eight years to get this to work’ he said, adding they were amazed because ‘it worked really, really well’.
This extends work conducted in his laboratory over more than a decade in which he has, for example, created an ‘orthogonal ribosome’ to work alongside the real thing while leaving the original ribosomes to take care of key cellular functions. It should now be possible to engineer the orthogonal ribosome to assemble the new building blocks more efficiently into proteins.
The ability to add new building blocks to proteins could, for example, make a protein-based drug resistant to enzymes that destroy proteins – which can limit their use in the body – thereby provide a foundation for accelerating the discovery of new therapeutics. The new building blocks can also lock a protein into the right shape, which is critical for ensuring that they work properly in the body.
ACCELERATING EVOLUTION
The team has extended the toolkit of life in other ways. Today, in the journal Science, they report a way to speed up the process that has shaped all life on Earth: they can now evolve useful genes in E. coli using ‘orthogonal replication’, overcoming a key hurdle to turbocharging evolution in the laboratory.
In natural evolution mutations introduced, when copying the genomic DNA of an organism, thrive in the environment if they provide an advantage to the organism’s offspring. However, most mutations are deleterious and therefore organisms need to limit their rate of mutation, and therefore the rate at which they can evolve, to avoid extinction. As a result, they tend to acquire new functions very slowly.
One way to do accelerate evolution in the laboratory is to put a gene of interest in a virus, which can be mutated, but it is limited to small genes and enhancing properties linked with infectivity. Another approach has been tried in yeast, but again there are limitations.
Now Rongzhen Tian and other members of Chin’s team have found a way to accelerate the rate at which entire organisms can acquire new functions in targeted genes without creating deleterious effects, again using E. coli. ‘All you have to do is grow these cells and they’re continuously mutating the DNA,’ Chin said. ‘In this way you can select for properties that are important to the cell that you couldn’t really select for with a test-tube methods.’
To achieve this they introduced DNA, which is not copied along with the organism’s genome, into a cell. They also took an enzyme called DNA polymerase, which is normally used to copy the DNA of a virus and created an ‘orthogonal version’ to selectively copy and rapidly mutate the newly introduced DNA, without affecting the organism’s own DNA.

By creating a second DNA copying system in cells, the Chin lab could massively accelerate the evolution of a new function in the introduced DNA sequences without introducing deleterious mutations. ‘The process is continuous and all you have to do is grow the cells,’ he said. In a few millilitres of liquid, it is possible to sample a trillion variations on one genetic theme, overcoming what is called the ‘critical mutation rate’.
In one demonstration, they showed how to increase the brightness of a cellular protein, called GFP, by 1000-fold, and how to create resistance to an antibiotic, to provide key insights that could help design drugs to curb the rise of superbugs. Other applications could be to rapidly evolve the ability of organisms to absorb the greenhouse gas carbon dioxide, for example, or to rapidly evolve antiviral drugs in a pandemic. And, of course, it can also be used to evolve proteins that contain his extended repertoire of amino acids.
TOOLKIT OF SYNTHETIC LIFE
In previous work, Chin’s team showed how to create the entire genetic recipe, or genome, of an organism such as a bacterium as a continuous process, using a tool developed by his team called CONEXER, which can write half a million ‘letters’ of genetic code in a few days. Another technology, called BASIS, allows his laboratory to grow big fragments – up a million genetic letters – of human DNA in E. coli.
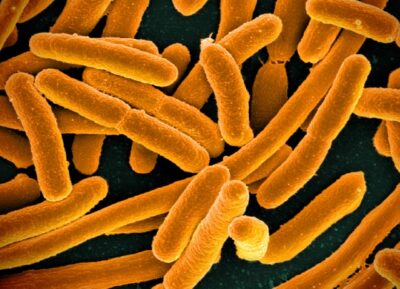
In this way, the organism can be used to handle large pieces of synthetic DNA from a range of organisms to create the entire genetic code of an organism such as a bacterium from scratch. They also used this to show how to compress the genetic code of E. coli, so it was more efficient than the one found in nature.
Aware of the unease about engineering novel lifeforms, he added that these novel organisms can only grow in laboratory conditions, so they are unable to reproduce in the wild.